|
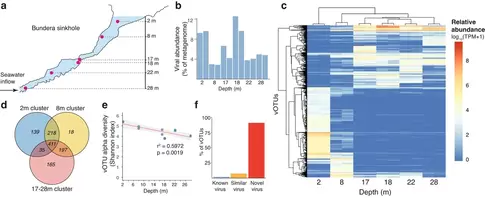
Timothy M. Ghaly, Amaranta Focardi, Liam D. H. Elbourne, Brodie Sutcliffe, William F. Humphreys, Paul R. Jaschke, Sasha G. Tetu and Ian T. Paulsen. (2024). Exploring virus-host-environment interactions in a chemotrophic-based underground estuary. Environmental Microbiome. DOI: https://doi.org/10.1186/s40793-024-00549-6
|
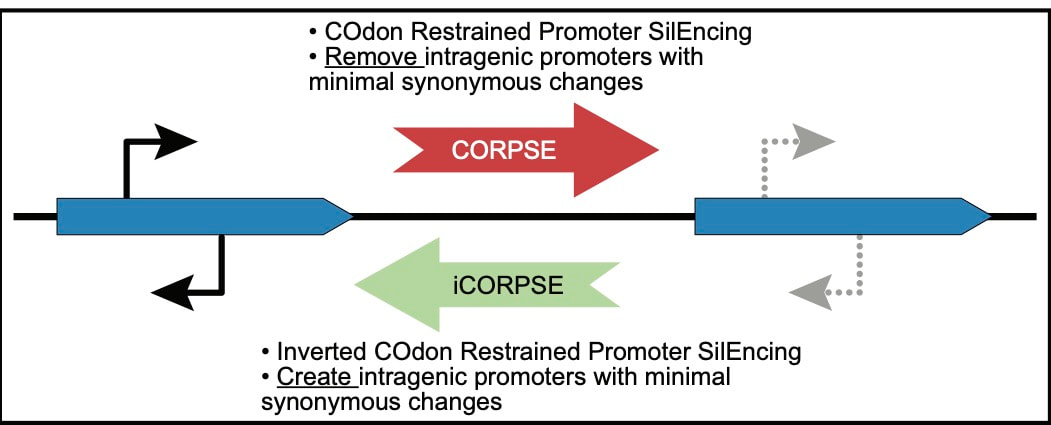
Ellina Trofimova, Dominic Y. Logel, and Paul R Jaschke. (2024). An Improved Method for Eliminating or Creating Intragenic Bacterial Promoters. In: Braman, JK (ed). Synthetic Biology: Methods and Protocols. Methods in Molecular Biology, vol. 2760. Humana, New York, NY. DOI: doi.org/10.1007/978-1-0716-3658-9_12
Pamela R. Tsoumbris, Russel M. Vincent, and Paul R Jaschke. (2024). Phage biocontainment using dependence on the amber initiator tRNA. (In Preparation).
Hannah X Zhu, Bradley W Wright, Dominic Y Logel, Patrick Needham, Kevin Yehl, Mark P Molloy, Paul R Jaschke. (2024). IbpAB small heat shock proteins are not host factors for bacteriophage φX174 replication. (In Preparation).
Shahla Asgharzadeh Kangachar, Dominic Y Logel, Ellina Trofimova, Hannah X Zhu, Julian Zaugg, Mark A Schembri, Karen D Weynberg, Paul R Jaschke. (2024). Discovery and characterisation of new phage targeting uropathogenic Escherichia coli. bioRxiv. DOI: doi.org/10.1101/2024.01.12.575291 (Under Review)
Ellina Trofimova, Shahla Asgharzadeh Kangachar, Karen D Weynberg, Robert Willows, and Paul R Jaschke. (2023). A bacterial genome assembly and annotation laboratory using a virtual machine. Biochemistry and Molecular Biology Education. DOI: doi.org/10.1002/bmb.21720
Dominic Y Logel, Paul R Jaschke (2023). Creating De Novo Overlapped Genes. In: Selvarajoo, K. (eds) Computational Biology and Machine Learning for Metabolic Engineering and Synthetic Biology. Methods in Molecular Biology, vol 2553. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2617-7_6
Article also on bioRxiv. DOI: https://doi.org/10.1101/2022.02.15.480625
Link to virtual machine download (warning! 19GB file).
Article also on bioRxiv. DOI: https://doi.org/10.1101/2022.02.15.480625
Link to virtual machine download (warning! 19GB file).
|
Bradley W Wright, Mark P Molloy, and Paul R Jaschke. (2022). Overlapping genes in natural and engineered genomes. Nature Reviews Genetics. rdcu.be/cySvX. Published Online: 5 October 2021
Abstract |
|
|
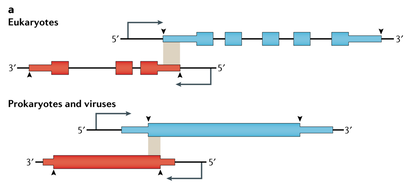
Dominic Y. Logel, Ellina Trofimova, and Paul R Jaschke. (2021). A Codon Restrained Method for Both Eliminating and Creating Intragenic Bacterial Promoters. ACS Syn. Bio. DOI: 10.1021/acssynbio.1c00359
Abstract |
Andras Hutvagner, Dominic Scopelliti, Fiona Whelan, and Paul R Jaschke. (2021). Orthogonal translation using the non-canonical initiator-tRNA(AAC) alters protein sequence and stability in vivo. bioRxiv. DOI: 10.1101/2021.05.25.445580
Abstract
Abstract
Bradley W Wright, Dominic Y Logel, Mehdi Mirzai, Dana Pascovici, Mark P Molloy, and Paul R Jaschke. (2020). Proteomic and transcriptomic analysis of Microviridae φXI74 infection reveals broad up-regulation of host membrane damage and heat shock responses. mSystems. DOI: 10.1128/mSystems.00046-21
Abstract
Abstract
|
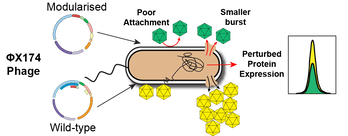
Bradley W Wright, Juanfang Ruan, Mark P Molloy, Paul R Jaschke. (2020). Genome modularization reveals overlapped gene topology is necessary for efficient viral reproduction. ACS Synthetic Biology. DOI: 10.1021/acssynbio.0c00323
Abstract Supplementary Information |
|
Dominic Y Logel and Paul R Jaschke. (2020). A high-resolution map of bacteriophage ϕX174 transcription. Virology. 547: 47-56. DOI: 10.1016/j.virol.2020.05.008
Abstract
Abstract
Paul R Jaschke. (2019). Simulated sandwich enzyme‐linked immunosorbent assay for a cost‐effective investigation of natural and engineered cellular signaling pathways. Biochemistry and Molecular Biology Education. 18 September 2019. DOI: https://doi.org/10.1002/bmb.21304.
Abstract PDF
Abstract PDF
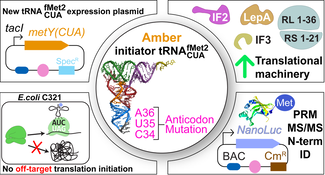
Russel M. Vincent, Pandelitsa F. Yiasemides, Paul R Jaschke. (2019). An orthogonal amber initiator tRNA functions similarly across diverse Escherichia coli laboratory strains.
ScienceMatters. 1 May 2019. DOI: 10.19185/matters.201904000009
Abstract PDF
ScienceMatters. 1 May 2019. DOI: 10.19185/matters.201904000009
Abstract PDF
|
NAR BREAKTHROUGH ARTICLE
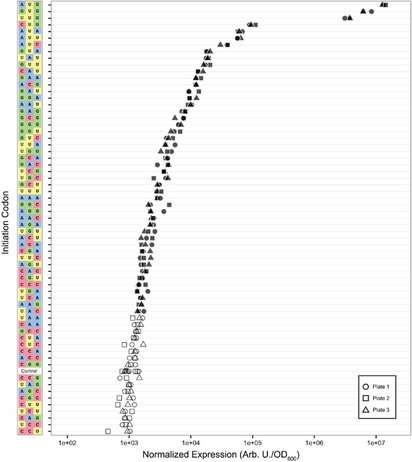
Ariel Hecht*, Jeff Glasgow*, Paul R. Jaschke*, Lukmaan Bawazer*, Matthew S. Munson, Jennifer Cochran, Drew Endy, Marc Salit. (2017). Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Research. 45 (7): 3615-3626. DOI: 10.1093/nar/gkx070 *Authors contributed equally to the paper as first authors. |
|
|
Figure 2. Translation initiation from all 64 codons. Normalized per-cell fluorescence measured from three replicate cultures, grown in LB and resuspended in PBS before measurement, with each of the 64 codons as the start codon in the GFP coding sequence. Shapes represent the replicate plate number, and filled shapes represent GFP expression significantly greater (adjusted P < 0.05) than the non-expressing control (Control) as determined by Dunnett’s test.
|
|
*Maxwell Bates, *Aaron Berliner, Joe Lachoff, Paul R. Jaschke, Eli Groban. (2016). Wet Lab Accelerator: A Web-Based Application Democratizing Laboratory Automation for Synthetic Biology. ACS Synthetic Biology. (2017), 6 (1), pp 167–171. DOI: 10.1021/acssynbio.6b00108
Abstract PDF *Authors contributed equally to this work |
|
|
Ariel Hecht*, Jeff Glasgow*, Paul R. Jaschke*, Lukmaan Bawazer*, Matthew S. Munson, Jennifer Cochran, Drew Endy, Marc Salit. (2016). Measurements of translation initiation from all 64 codons in E. coli. bioRxiv.
*Authors contributed equally to this work Link |
|
Hessel A, Quinn J, Jaschke PR. Sequence of Synthetic øX174 Bacteriophage. Museum of Modern Art (MoMA). Accessioned Item. New York, USA (Invited Exhibit)
Hessel A, Quinn J, Jaschke PR. Synthetic øX174 Bacteriophage. Design and Violence Exhibit. Museum of Modern Art (MoMA). New York, USA (Invited Exhibit).
Link
Link
Jaschke PR, Ju Lu, Widya Mulyasasmita, and Luke J. Lee. (2013). Incyte Pharmaceuticals Is Primed For A Run. Seeking Alpha.
Link
Link
|
Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D. (2012). A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast. Virology. (Cover).
DOI: 10.1016/j.virol.2012.09.020. Abstract PDF Refactored øX174.1f genome sequence: JX913857 |
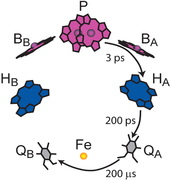
Neupane B, Jaschke P, Saer R, Beatty JT, Reppert M, Jankowiak R. (2012). Electron Transfer in Rhodobacter sphaeroides Reaction Centers Containing Zn-Bacteriochlorophylls: A Hole Burning Study. The Journal of Physical Chemistry B. Mar 15; 116(10): 3457-3466. DOI: 10.1021/jp300304r
Abstract PDF
Abstract PDF
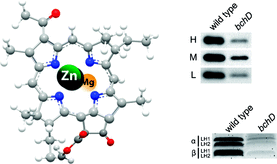
Jaschke PR, Hardjasa A, Digby E, Hunter CN, Beatty JT. (2011). A bchD (Mg-chelatase) mutant of Rhodobacter sphaeroides synthesizes zinc-bacteriochlorophyll through a novel zinc-containing intermediates. The Journal of Biological Chemistry. Jun 10;286(23):20313-22. DOI: 10.1074/jbc.M110.212605
Abstract PDF
Abstract PDF
Jaschke PR, Saer RG, Noll S, Beatty JT. (2011). Modification of the genome of Rhodobacter sphaeroides and construction of synthetic operons. Methods in Enzymology. 497:519-38. DOI: 10.1016/B978-0-12-385075-1.00023-8
Abstract PDF
Abstract PDF
Jaschke PR. (2010). Discovery and characterization of a new zinc-bacteriochlorophyll biosynthetic pathway and photosystem in a magnesium-chelatase mutant. PhD Thesis. University of British Columbia. Abstract PDF
|
Lin S, Jaschke PR, Wang H, Paddock M, Tufts A, Allen JP, Rosell FI, Mauk GA, Woodbury NW, Beatty JT. (2009). Electron transfer in the Rhodobacter sphaeroides reaction center assembled with zinc bacteriochlorophyll. Proceedings of the National Academy of Sciences of the USA. 106(21): 8537-42.
DOI: 10.1073/pnas.0812719106 Abstract PDF |
Jaschke PR, LeBlanc HN, Lang AS, Beatty JT. (2008). The PucC protein of Rhodobacter capsulatus mitigates an inhibitory effect of light-harvesting 2 alpha and beta proteins on light-harvesting complex 1. Photosynthesis Research. 95(2-3): 279-84. DOI: 10.1007/s11120-007-9258-x
Abstract PDF
Abstract PDF